A Brief Tutorial on Biodatascreen
By Roseli Pellens, Thomas Haevermans, Visotheary Ung, Antoine Bisch, and Frederic Legendre
This tutorial gives basic information regarding how to use
BIODATASCREEN pipeline developed by ISYEB, Institut de
Systématique, Evolution, Biodiversité. This project was funded by
CNRS - INEE
Table of Contents
1. Upload
Your files must strictly conform to the .csv format.
Format
conversion can easily be done in Excel or any spreadsheet software
by selecting the correct saving option (using UTF-8 encoding).
Separator
Select the separator used in your .csv file. If sure, perform a quick visual check with a simple text editor. After selecting the correct separator, you can reach the mapping step (as in Figure 1). If not, please re-check the separator.

Figure 1. A screenshot from biodatascreen's pipeline page: Mapping
Mapping to Darwin Core format
This step ensures that the columns’ contents are correctly read.
This mapping tool recognizes column headers when they match
a Darwin Core term (case insensitive). If you are dealing with a
table including a large number of columns it can be more practical
to rename them directly in your file before uploading.
For
columns not recognized automatically, you need to open the window
as in the figure above and select the term corresponding to the
contents of your column.
The only mandatory columns to run the pipeline are
decimalLatitude, decimalLongitude and countryCode. You must
ascertain that these three columns are correctly mapped from your
original file. Coordinates must be in decimal degrees – no
conversion tool is offered.
Only the successfully mapped columns (those you mapped, or
those recognized automatically) will be present in your output
files. Any other column will be discarded.
For more information go to Darwin Core
http://rs.tdwg.org/dwc/terms/index.htm
or
http://rs.tdwg.org/dwc/terms/history/dwctoabcd/index.htm
If you receive a message like “something is wrong”
at this step, go back to your input file and check
- the presence of columns without header – they should be deleted as each column must have a unique header
- the presence of columns with duplicate headers – be sure that each column is named differently – case insensitive
- the presence of white space(s) at the beginning of the column(s) titles – column(s) with such header will not be read, and it can lead to errors, wrong results or no results
- the way decimalLatitude and decimalLongitude are spelled – they must be in US notation; for example, -21.1333, 165.1427 for a place in New Caledonia.
sometimes excel makes .csv files that are not fully compliant with utf-8 code, (particularly excel in mac) and this will block the pipeline parser.
If you still receive the message “something is wrong” after having checked and corrected the points above, please try to use another way to save your csv file. You can use texteditor or notebook to check the presence of unrecognized characters. Letters with diacritics such, for instance, those used in several Latin languages, can be a source of unrecognized characters.
See more about encoding to utf-8 with excel here.
Uploading several files
Several files can be uploaded in a single run, but you must select
the separators and map towards Darwin Core one file at a time.
Once a file has been added, you cannot go backwards and map or
change the mapping in the previous ones. If you have a problem
with a file that is already mapped, the only way to fix it is to
delete the file and then upload it again, choose the separator and
map to Darwin Core again.
To avoid this kind of problem, be
sure to determine the columns you want to map before you start.
The output files are a consolidation of the input files.
So, if you map different columns for different files, these
columns will have data only in the part concerning the file you
mapped them from.
What to do if your file has a column without any
match among the 250 column terms from Darwin Core?
The
easiest solution is to search for an empty comment term (one not
used by your file) and name your column alike.
2. Pipeline settings
The pipeline includes one mandatory setting and four optional
ones.
Beware to choose carefully the options you want.
The mandatory step is the Geographic Coordinates Validation.
Geographic Coordinates Validation
At this step, the pipeline checks for
- ISO2 – for the presence of ISO2 code for each occurrence;
- Presence of two valid coordinates (decimalLatitude and decimalLongitude)
- Congruence between geographic coordinates and country code (ISO2 code)
- HasGeospatialIssue – Gbif parameter: http://www.gbif.org/developer/occurrence#p_hasGeospatialIssue
– the Gbif parameter hasGeospatialIssue indicates the existence
of a problem concerning the spatial information of an occurrence.
When hasGeospatialIssue= true one should take a closer look at
the occurrence, instead of using it automatically.
In this pipeline only records without spatial issues are kept, i.e., those for which hasGeospatialIssue = false (this is not case sensitive, so False or FALSE or false can be used).
Note: when the information hasGeospatialIssue is absent, the pipeline assumes that hasGeospatialIssue = false
The geographic coordinates system used is WGS84
Using a different coordinate system does not block the pipeline, but the results may not be accurate.
As you know, when systems change, so does the relative position of places on maps, and it may affect primarily those occurrences that are on the edge of your map due to different projection systems artifacts. The most obvious expected effect of using a different projection system will be discarding good occurrences on one side of the map and including wrong occurrences on the other.
Filtering mask dependent errors
This tool verifies if occurrences are consistently included in a
raster surface. More information on rasters here.
For this, you can use our default
mask or one of your own.
The default mask is designed to
filter data that are in land cells, i.e., it is intended to
discard points falling in water bodies and in oceans. The default
mask is the commonly used file BIO12 (= Annual Precipitation) at
the highest resolution (30 arc-seconds ~1 km) from WorldClim (http://www.worldclim.org/current).
We opted for this layer because it covers and returns values for
the greatest majority of the inland cells.
This pipeline
works with .bil files for which it is convenient to add the .bil
and the .hdr (headers) files, even though only the .bil file is
required.
By using your own resources you can also use this function
to filter your data for a limited region. For instance, when
working in Madagascar you can use a mask to keep only the
occurrences inside its boundaries. But, please pay attention on
the resolution of the raster you use. As you can see in the figure
above, the finer the grain, the more accurate is the solution. In
other words, a raster with high resolution reduces the possibility
of deleting good occurrences or including bad occurrences at the
boundaries of a selected area.
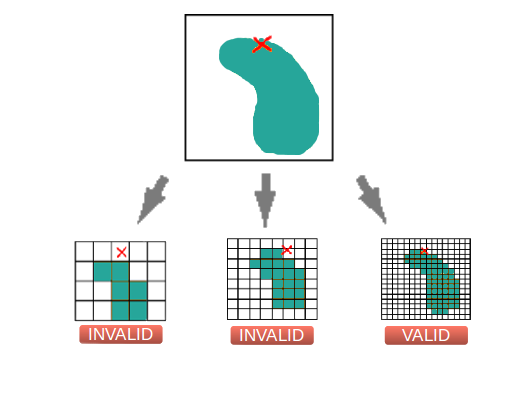 Figure 2. An illustration
showing how the grain of the grid may influence the accuracy of
the results.
Figure 2. An illustration
showing how the grain of the grid may influence the accuracy of
the results. Set TDWG4 Code
The Taxonomic Database Working Group (TDWG) system provides clear
definitions and codes for recording plant distributions at four
scales or levels, from "botanical continents" – level 1 – down to
parts of large countries – level 4. Several databases have this
information available for plants, but you can integrate this level
to any organisms you want.
At this step the pipeline checks
whether the coordinates fall within a known polygon from the TDWG
scheme for recording plant distribution*. If the information is
already present in your file the pipeline checks whether the point
falls in the correct polygon, otherwise the pipeline will fill in
the data with the corresponding polygon.
If this option is
selected, the output file will contain TDWG level 4 polygon
information for all records.
*Brummitt, R. K. Plant
taxonomic databases standards N°2 ed. 2. World geographical
scheme for recording plant distributions, ed. 2. (2001)
Filter by establishment status
EstablishmentMeans is a Gbif parameter that characterizes the way
the species arrived in a locality. Possible values are:
Introduced, Invasive, Managed, Native, Naturalised, Uncertain.
This function gives the option of selecting data based on a column
providing this information, i.e. establishmentMeans. The final
result of the pipeline will contain only the occurrences for the
selected categories.
To use this tool you need:
- to provide a file with a establishmentMeans column
- to ensure that the column contains the information you
want for each occurrence;
Lines without information about the establishmentMeans will be discarded unless you choose the option “other”.
Note: Several values may be selected at a time.
The pipeline does not add any information, it works by selecting the lines with the filter you enforce.
Reconciling data
This tool works by comparing names in the ‘ScientificName’ column
with names in a selected database. Two databases are offered:
Kew.org database, which only contains botanical names; and
Gbif.org database, which covers more organisms.
The reconciliation is made on the ‘ScientificName’ column
so be sure this column is available in your file.
This option requires two steps, first selection of the
option, then selection of the reconciliation service. Make sure to
select both.
Note: Only names with a 100% match are retained. If
your file contains names with different spellings they will be
discarded alongside those with spelling errors.
All
occurrences without 100% match can be found in the output file
no-reconciliation.csv, so you can identify and correct them using
less stringent match conditions (with OpenRefine for
example).
3. Obtaining your results
Note for teachers:
You are very welcome to use this
pipeline for your courses and trainings. The more it is used the
worthier it is to have made it, but please be aware of the
webserver's limits, which may be not capable to support hundreds
of users at the same time.
We recommend limiting the size of
the input files to up to 15,000 lines. This way you can be sure to
have your results in a short time-frame and you can run several
pipelines at the same time.
When launching several pipelines
at a same time please ask for obtaining the results by e-mail.
Receiving your Results
There are two ways to get your results. You can either get them
directly on line, then wait until the analysis is entirely
completed, or receive them by e-mail.
Unless you are using a
small amount of data – maximum 15,000 lines – we recommend the
email option (please make sure to provide a correct e-mail
address).
The output takes the form of a set of files: one file
mapping_dwc.csv; different files with the occurrences possibly
excluded at each step, one with the filtered data, named
clean_table.csv, and one log file containing all pipeline’s steps
(see table bellow)
Note: Only one file is generated at each step,
whatever the number of input files.
A zip file containing
all these outputs is also available.
Contents of the output files generated by the pipeline.
| File name | Contents |
|---|---|
| mapping_dwc.csv | Mapped data of input file(s) with darwinCore headers |
| discarded_iso2.csv | Occurrences without or with unknown ISO2 code |
| discarded_coordinates.csv | Occurrences with latitude outside [-90,90] degree or longitude outside [-180,180] degree |
| discarded_geospatialIssues.csv | Occurrences with geospatialIssues set to true |
| discarded_polygon.csv | Occurrences with coordinates that do not correspond to the country code indicated |
| discarded_raster.csv | Occurrences not compliant with any mask(s) provided |
| cells_proba_raster.csv | Details of all the occurrences with for each the result for each masks |
| no_establishment_means.csv | Occurrences not corresponding to the establishment mean(s) chosen |
| no_reconciliation.csv | Occurrences for which the scientific name did not match at 100% to one in the database you chose |
| clean_table.csv | This is the file with occurrences that passed all requested filters |
| execution_log | A text file with requested pipeline’ steps name followed by true (if the step was correctly done), or false (if an error occurred) |
| all_files.zip | Archive containing all output files produced by the pipeline |